Log media and objects
12 minute read
We support images, video, audio, and more. Log rich media to explore your results and visually compare your runs, models, and datasets. Read on for examples and how-to guides.
Pre-requisites
In order to log media objects with the W&B SDK, you may need to install additional dependencies. You can install these dependencies by running the following command:
pip install wandb[media]
Images
Log images to track inputs, outputs, filter weights, activations, and more.

Images can be logged directly from NumPy arrays, as PIL images, or from the filesystem.
Each time you log images from a step, we save them to show in the UI. Expand the image panel, and use the step slider to look at images from different steps. This makes it easy to compare how a model’s output changes during training.
Provide arrays directly when constructing images manually, such as by using make_grid from torchvision.
Arrays are converted to png using Pillow.
images = wandb.Image(image_array, caption="Top: Output, Bottom: Input")
wandb.log({"examples": images})
We assume the image is gray scale if the last dimension is 1, RGB if it’s 3, and RGBA if it’s 4. If the array contains floats, we convert them to integers between 0 and 255. If you want to normalize your images differently, you can specify the mode manually or just supply a PIL.Image, as described in the “Logging PIL Images” tab of this panel.
For full control over the conversion of arrays to images, construct the PIL.Image yourself and provide it directly.
images = [PIL.Image.fromarray(image) for image in image_array]
wandb.log({"examples": [wandb.Image(image) for image in images]})
For even more control, create images however you like, save them to disk, and provide a filepath.
im = PIL.fromarray(...)
rgb_im = im.convert("RGB")
rgb_im.save("myimage.jpg")
wandb.log({"example": wandb.Image("myimage.jpg")})
Image overlays
Log semantic segmentation masks and interact with them (altering opacity, viewing changes over time, and more) via the W&B UI.

To log an overlay, provide a dictionary with the following keys and values to the masks keyword argument of wandb.Image:
- one of two keys representing the image mask:
"mask_data": a 2D NumPy array containing an integer class label for each pixel"path": (string) a path to a saved image mask file
"class_labels": (optional) a dictionary mapping the integer class labels in the image mask to their readable class names
To log multiple masks, log a mask dictionary with multiple keys, as in the code snippet below.
mask_data = np.array([[1, 2, 2, ..., 2, 2, 1], ...])
class_labels = {1: "tree", 2: "car", 3: "road"}
mask_img = wandb.Image(
image,
masks={
"predictions": {"mask_data": mask_data, "class_labels": class_labels},
"ground_truth": {
# ...
},
# ...
},
)
Segmentation masks for a key are defined at each step (each call to wandb.log()).
- If steps provide different values for the same mask key, only the most recent value for the key is applied to the image.
- If steps provide different mask keys, all values for each key are shown, but only those defined in the step being viewed are applied to the image. Toggling the visibility of masks not defined in the step do not change the image.
Log bounding boxes with images, and use filters and toggles to dynamically visualize different sets of boxes in the UI.
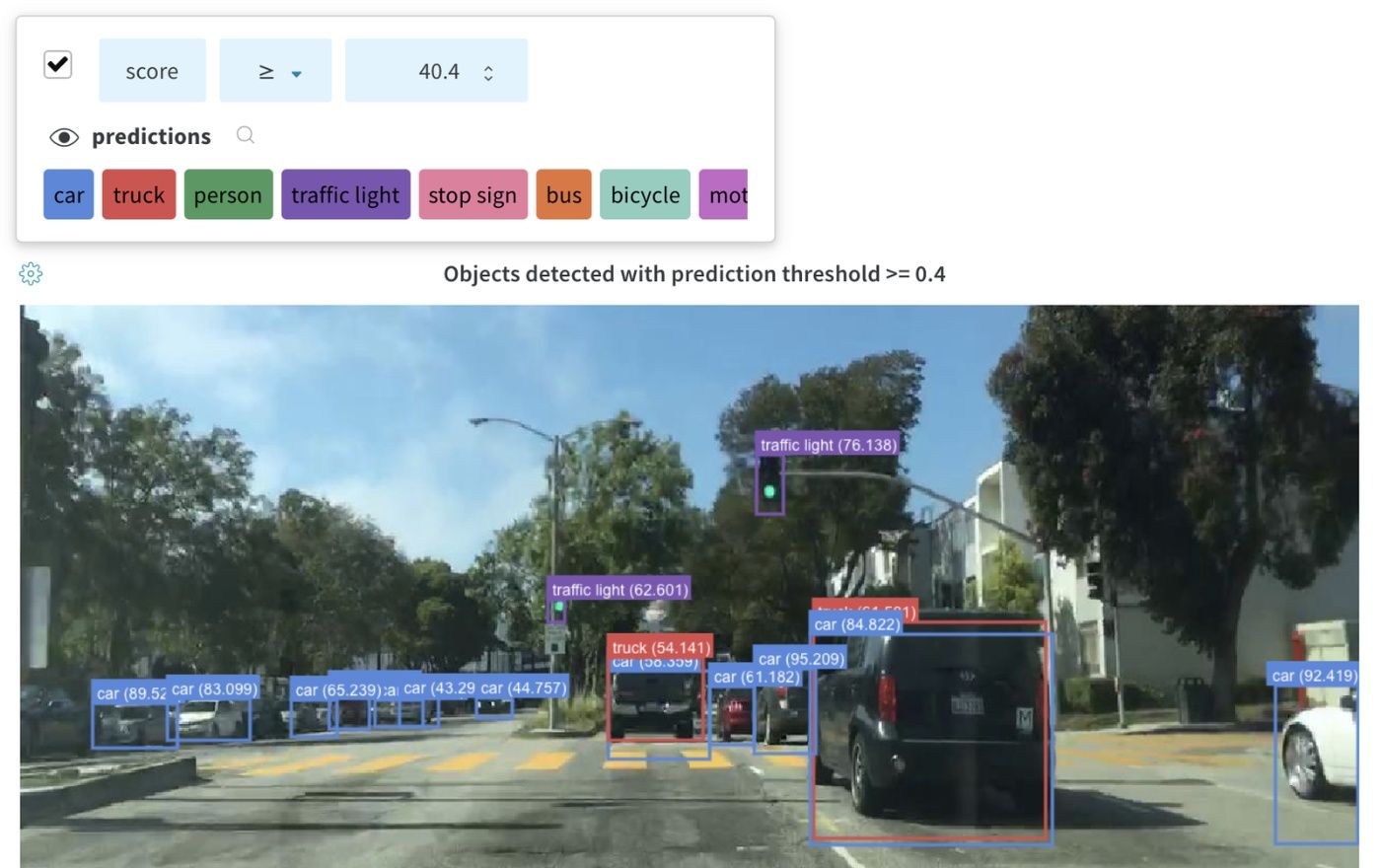
To log a bounding box, you’ll need to provide a dictionary with the following keys and values to the boxes keyword argument of wandb.Image:
box_data: a list of dictionaries, one for each box. The box dictionary format is described below.position: a dictionary representing the position and size of the box in one of two formats, as described below. Boxes need not all use the same format.- Option 1:
{"minX", "maxX", "minY", "maxY"}. Provide a set of coordinates defining the upper and lower bounds of each box dimension. - Option 2:
{"middle", "width", "height"}. Provide a set of coordinates specifying themiddlecoordinates as[x,y], andwidthandheightas scalars.
- Option 1:
class_id: an integer representing the class identity of the box. Seeclass_labelskey below.scores: a dictionary of string labels and numeric values for scores. Can be used for filtering boxes in the UI.domain: specify the units/format of the box coordinates. Set this to “pixel” if the box coordinates are expressed in pixel space, such as integers within the bounds of the image dimensions. By default, the domain is assumed to be a fraction/percentage of the image, expressed as a floating point number between 0 and 1.box_caption: (optional) a string to be displayed as the label text on this box
class_labels: (optional) A dictionary mappingclass_ids to strings. By default we will generate class labelsclass_0,class_1, etc.
Check out this example:
class_id_to_label = {
1: "car",
2: "road",
3: "building",
# ...
}
img = wandb.Image(
image,
boxes={
"predictions": {
"box_data": [
{
# one box expressed in the default relative/fractional domain
"position": {"minX": 0.1, "maxX": 0.2, "minY": 0.3, "maxY": 0.4},
"class_id": 2,
"box_caption": class_id_to_label[2],
"scores": {"acc": 0.1, "loss": 1.2},
# another box expressed in the pixel domain
# (for illustration purposes only, all boxes are likely
# to be in the same domain/format)
"position": {"middle": [150, 20], "width": 68, "height": 112},
"domain": "pixel",
"class_id": 3,
"box_caption": "a building",
"scores": {"acc": 0.5, "loss": 0.7},
# ...
# Log as many boxes an as needed
}
],
"class_labels": class_id_to_label,
},
# Log each meaningful group of boxes with a unique key name
"ground_truth": {
# ...
},
},
)
wandb.log({"driving_scene": img})
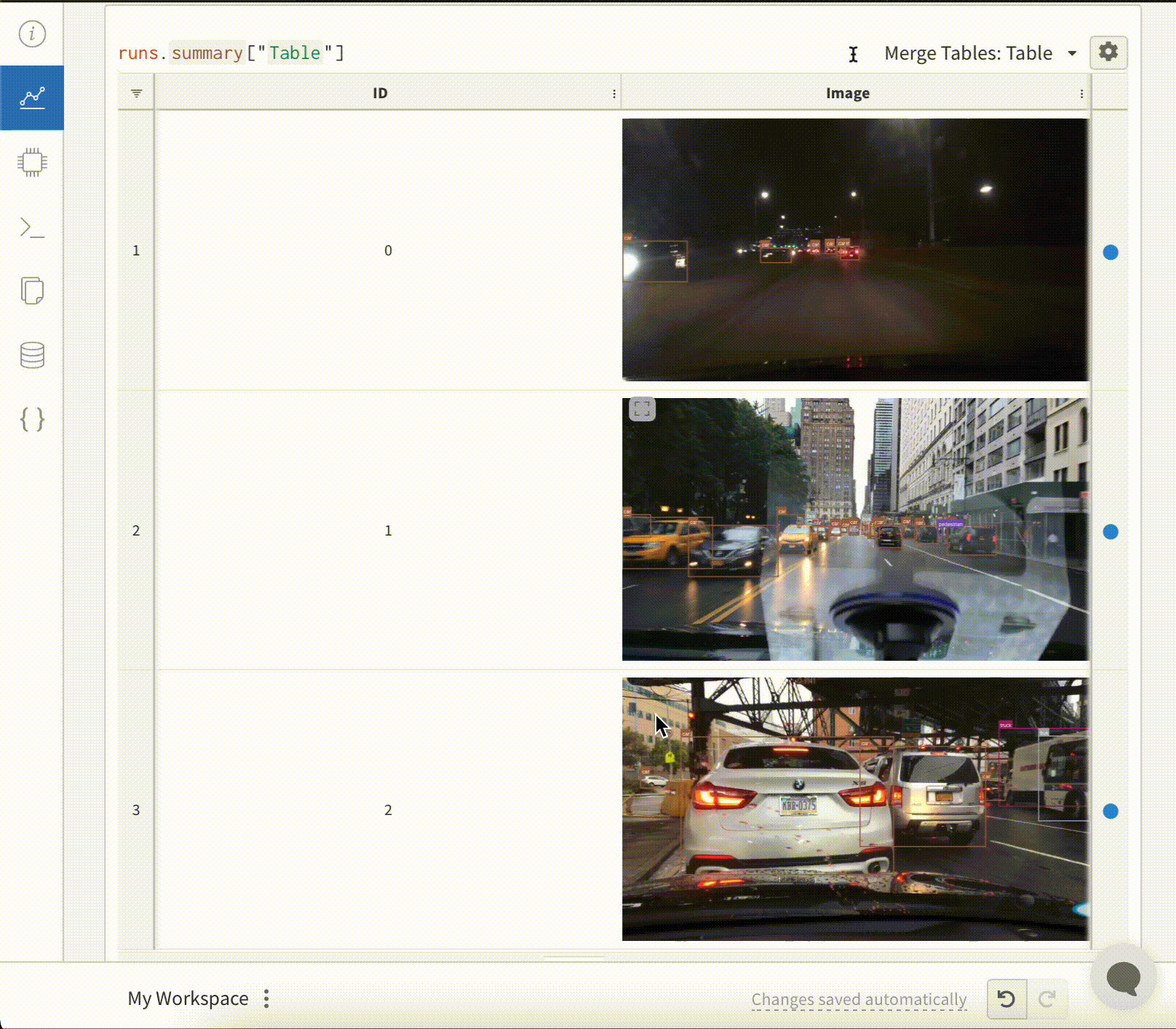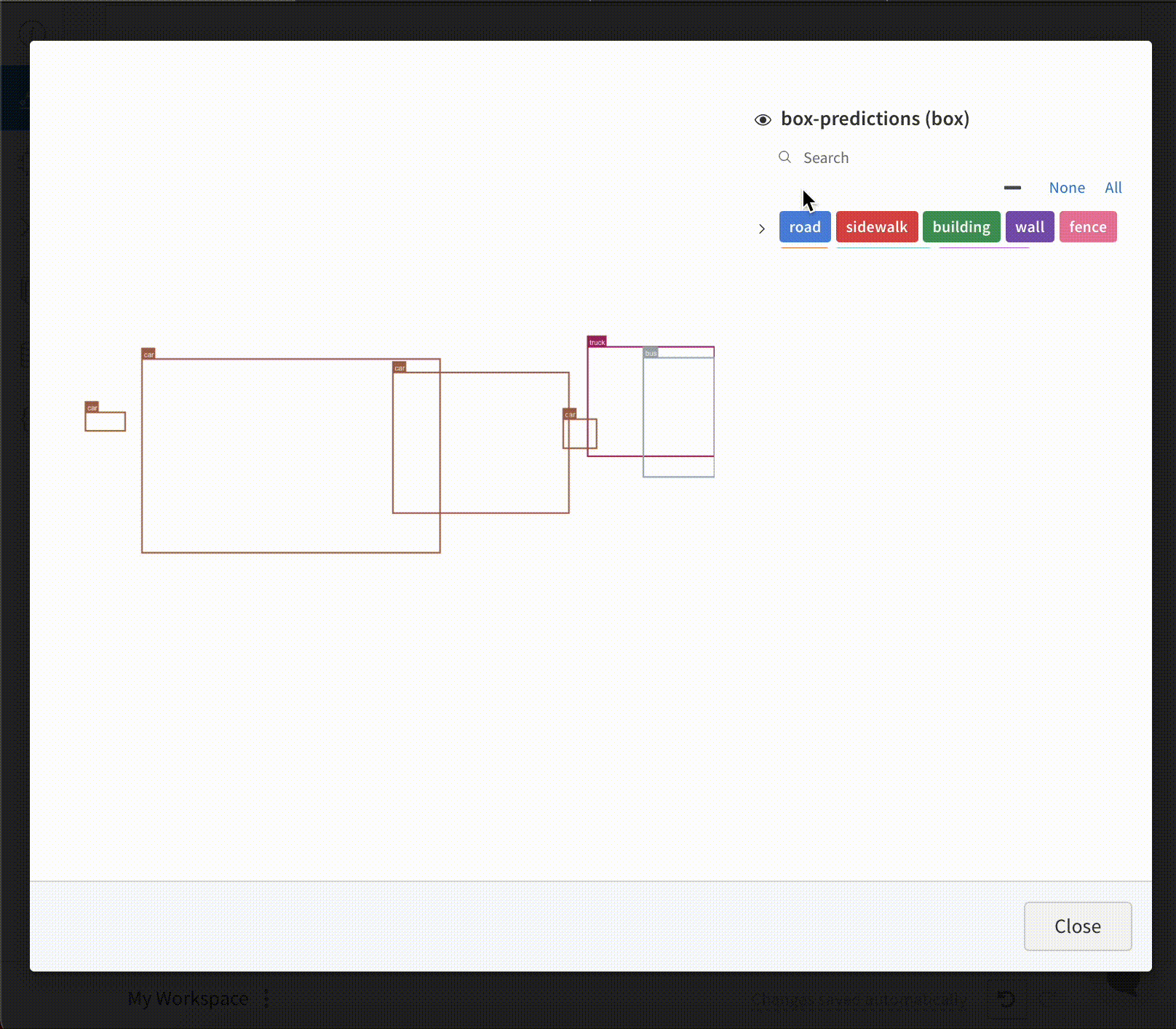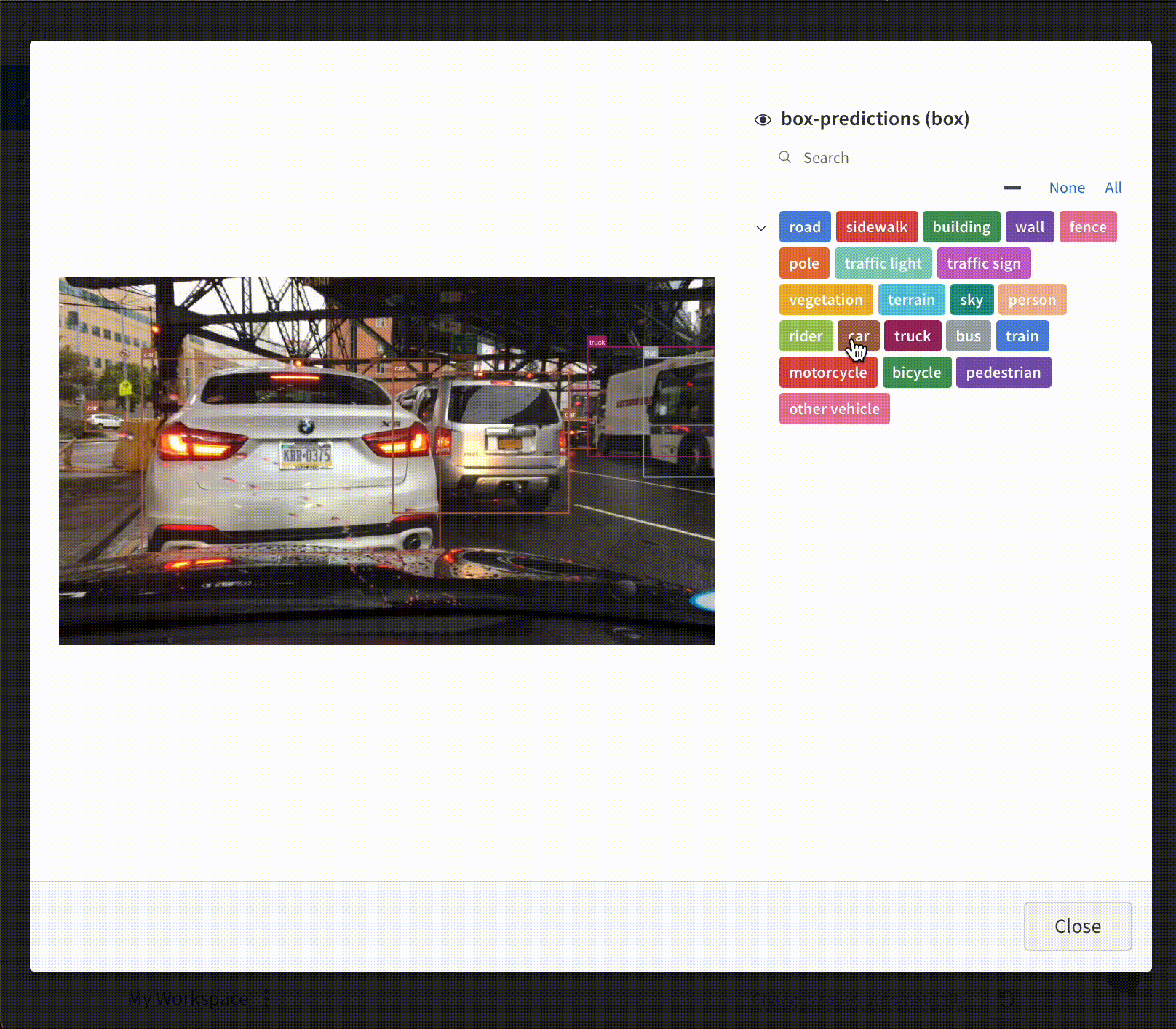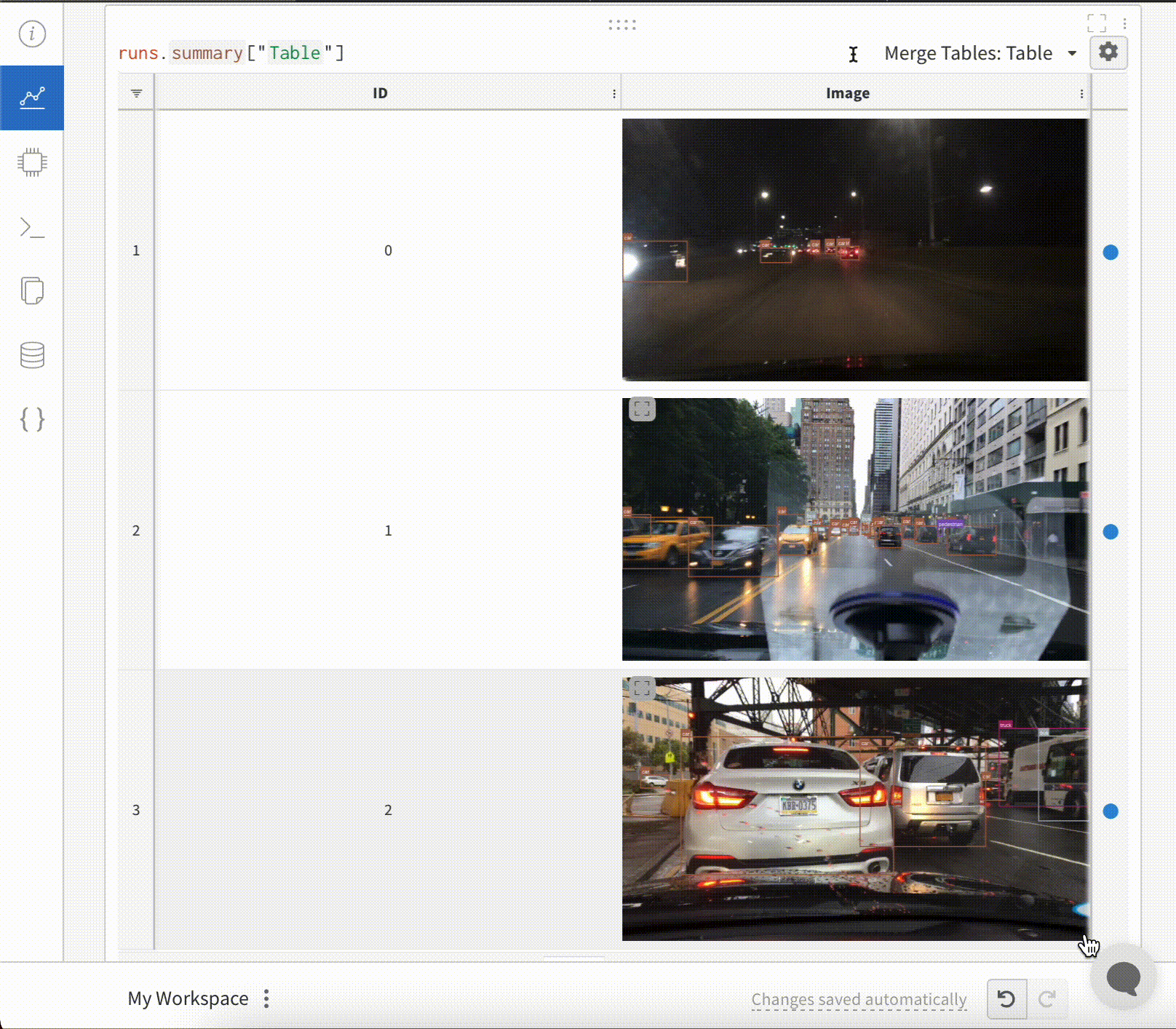
Image overlays in Tables
To log Segmentation Masks in tables, you will need to provide a wandb.Image object for each row in the table.
An example is provided in the Code snippet below:
table = wandb.Table(columns=["ID", "Image"])
for id, img, label in zip(ids, images, labels):
mask_img = wandb.Image(
img,
masks={
"prediction": {"mask_data": label, "class_labels": class_labels}
# ...
},
)
table.add_data(id, mask_img)
wandb.log({"Table": table})
To log Images with Bounding Boxes in tables, you will need to provide a wandb.Image object for each row in the table.
An example is provided in the code snippet below:
table = wandb.Table(columns=["ID", "Image"])
for id, img, boxes in zip(ids, images, boxes_set):
box_img = wandb.Image(
img,
boxes={
"prediction": {
"box_data": [
{
"position": {
"minX": box["minX"],
"minY": box["minY"],
"maxX": box["maxX"],
"maxY": box["maxY"],
},
"class_id": box["class_id"],
"box_caption": box["caption"],
"domain": "pixel",
}
for box in boxes
],
"class_labels": class_labels,
}
},
)
Histograms
If a sequence of numbers, such as a list, array, or tensor, is provided as the first argument, we will construct the histogram automatically by calling np.histogram. All arrays/tensors are flattened. You can use the optional num_bins keyword argument to override the default of 64 bins. The maximum number of bins supported is 512.
In the UI, histograms are plotted with the training step on the x-axis, the metric value on the y-axis, and the count represented by color, to ease comparison of histograms logged throughout training. See the “Histograms in Summary” tab of this panel for details on logging one-off histograms.
wandb.log({"gradients": wandb.Histogram(grads)})
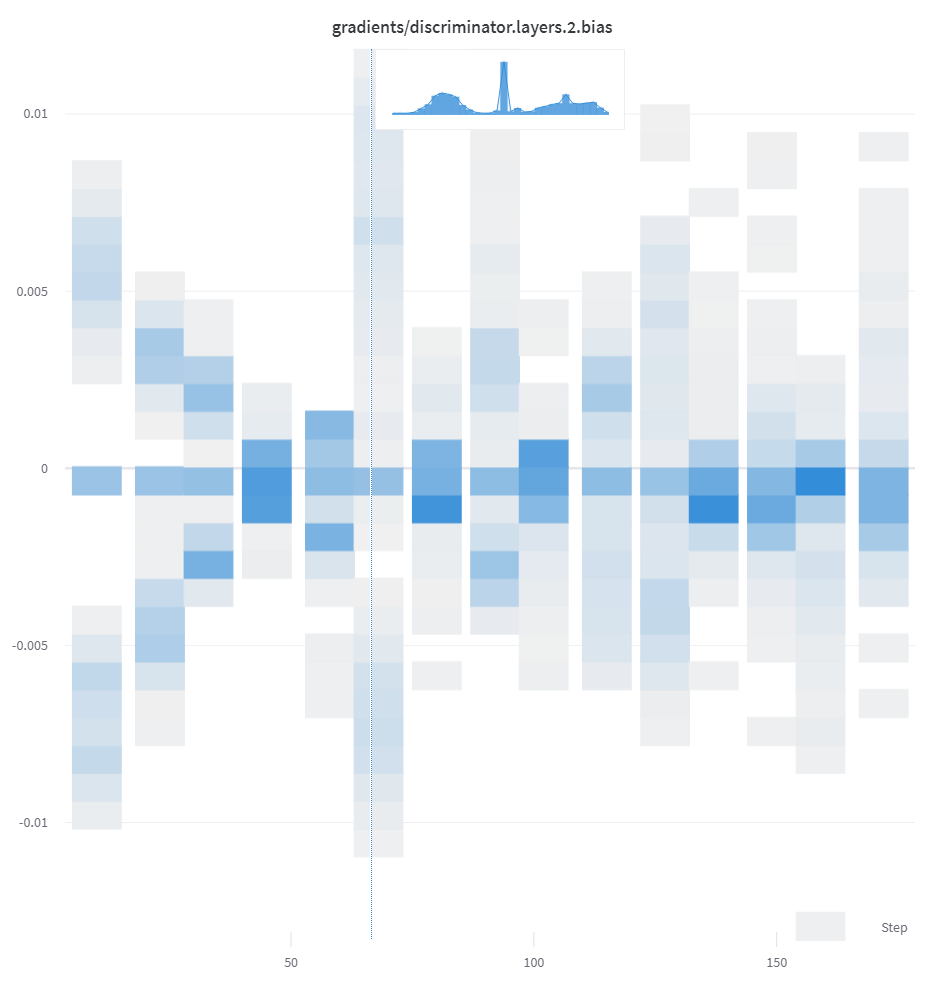
If you want more control, call np.histogram and pass the returned tuple to the np_histogram keyword argument.
np_hist_grads = np.histogram(grads, density=True, range=(0.0, 1.0))
wandb.log({"gradients": wandb.Histogram(np_hist_grads)})
If histograms are in your summary they will appear on the Overview tab of the Run Page. If they are in your history, we plot a heatmap of bins over time on the Charts tab.
3D visualizations
Log 3D point clouds and Lidar scenes with bounding boxes. Pass in a NumPy array containing coordinates and colors for the points to render.
point_cloud = np.array([[0, 0, 0, COLOR]])
wandb.log({"point_cloud": wandb.Object3D(point_cloud)})
NumPy array formats
Three different formats of NumPy arrays are supported for flexible color schemes.
[[x, y, z], ...]nx3[[x, y, z, c], ...]nx4| c is a categoryin the range[1, 14](Useful for segmentation)[[x, y, z, r, g, b], ...]nx6 | r,g,bare values in the range[0,255]for red, green, and blue color channels.
Python object
Using this schema, you can define a Python object and pass it in to the from_point_cloud method.
pointsis a NumPy array containing coordinates and colors for the points to render using the same formats as the simple point cloud renderer shown above.boxesis a NumPy array of python dictionaries with three attributes:corners- a list of eight cornerslabel- a string representing the label to be rendered on the box (Optional)color- rgb values representing the color of the boxscore- a numeric value that will be displayed on the bounding box that can be used to filter the bounding boxes shown (for example, to only show bounding boxes wherescore>0.75). (Optional)
typeis a string representing the scene type to render. Currently the only supported value islidar/beta
point_list = [
[
2566.571924017235, # x
746.7817289698219, # y
-15.269245470863748,# z
76.5, # red
127.5, # green
89.46617199365393 # blue
],
[ 2566.592983606823, 746.6791987335685, -15.275803826279521, 76.5, 127.5, 89.45471117247024 ],
[ 2566.616361739416, 746.4903185513501, -15.28628929674075, 76.5, 127.5, 89.41336375503832 ],
[ 2561.706014951675, 744.5349468458361, -14.877496818222781, 76.5, 127.5, 82.21868245418283 ],
[ 2561.5281847916694, 744.2546118233013, -14.867862032341005, 76.5, 127.5, 81.87824684536432 ],
[ 2561.3693562897465, 744.1804761656741, -14.854129178142523, 76.5, 127.5, 81.64137897587152 ],
[ 2561.6093071504515, 744.0287526628543, -14.882135189841177, 76.5, 127.5, 81.89871499537098 ],
# ... and so on
]
run.log({"my_first_point_cloud": wandb.Object3D.from_point_cloud(
points = point_list,
boxes = [{
"corners": [
[ 2601.2765123137915, 767.5669506323393, -17.816764802288663 ],
[ 2599.7259021588347, 769.0082337923552, -17.816764802288663 ],
[ 2599.7259021588347, 769.0082337923552, -19.66876480228866 ],
[ 2601.2765123137915, 767.5669506323393, -19.66876480228866 ],
[ 2604.8684867834395, 771.4313904894723, -17.816764802288663 ],
[ 2603.3178766284827, 772.8726736494882, -17.816764802288663 ],
[ 2603.3178766284827, 772.8726736494882, -19.66876480228866 ],
[ 2604.8684867834395, 771.4313904894723, -19.66876480228866 ]
],
"color": [0, 0, 255], # color in RGB of the bounding box
"label": "car", # string displayed on the bounding box
"score": 0.6 # numeric displayed on the bounding box
}],
vectors = [
{"start": [0, 0, 0], "end": [0.1, 0.2, 0.5], "color": [255, 0, 0]}, # color is optional
],
point_cloud_type = "lidar/beta",
)})
When viewing a point cloud, you can hold control and use the mouse to move around inside the space.
Point cloud files
You can use the from_file method to load in a JSON file full of point cloud data.
run.log({"my_cloud_from_file": wandb.Object3D.from_file(
"./my_point_cloud.pts.json"
)})
An example of how to format the point cloud data is shown below.
{
"boxes": [
{
"color": [
0,
255,
0
],
"score": 0.35,
"label": "My label",
"corners": [
[
2589.695869075582,
760.7400443552185,
-18.044831294622487
],
[
2590.719039645323,
762.3871153874499,
-18.044831294622487
],
[
2590.719039645323,
762.3871153874499,
-19.54083129462249
],
[
2589.695869075582,
760.7400443552185,
-19.54083129462249
],
[
2594.9666662674313,
757.4657929961453,
-18.044831294622487
],
[
2595.9898368371723,
759.1128640283766,
-18.044831294622487
],
[
2595.9898368371723,
759.1128640283766,
-19.54083129462249
],
[
2594.9666662674313,
757.4657929961453,
-19.54083129462249
]
]
}
],
"points": [
[
2566.571924017235,
746.7817289698219,
-15.269245470863748,
76.5,
127.5,
89.46617199365393
],
[
2566.592983606823,
746.6791987335685,
-15.275803826279521,
76.5,
127.5,
89.45471117247024
],
[
2566.616361739416,
746.4903185513501,
-15.28628929674075,
76.5,
127.5,
89.41336375503832
]
],
"type": "lidar/beta"
}
NumPy arrays
Using the same array formats defined above, you can use numpy arrays directly with the from_numpy method to define a point cloud.
run.log({"my_cloud_from_numpy_xyz": wandb.Object3D.from_numpy(
np.array(
[
[0.4, 1, 1.3], # x, y, z
[1, 1, 1],
[1.2, 1, 1.2]
]
)
)})
run.log({"my_cloud_from_numpy_cat": wandb.Object3D.from_numpy(
np.array(
[
[0.4, 1, 1.3, 1], # x, y, z, category
[1, 1, 1, 1],
[1.2, 1, 1.2, 12],
[1.2, 1, 1.3, 12],
[1.2, 1, 1.4, 12],
[1.2, 1, 1.5, 12],
[1.2, 1, 1.6, 11],
[1.2, 1, 1.7, 11],
]
)
)})
run.log({"my_cloud_from_numpy_rgb": wandb.Object3D.from_numpy(
np.array(
[
[0.4, 1, 1.3, 255, 0, 0], # x, y, z, r, g, b
[1, 1, 1, 0, 255, 0],
[1.2, 1, 1.3, 0, 255, 255],
[1.2, 1, 1.4, 0, 255, 255],
[1.2, 1, 1.5, 0, 0, 255],
[1.2, 1, 1.1, 0, 0, 255],
[1.2, 1, 0.9, 0, 0, 255],
]
)
)})
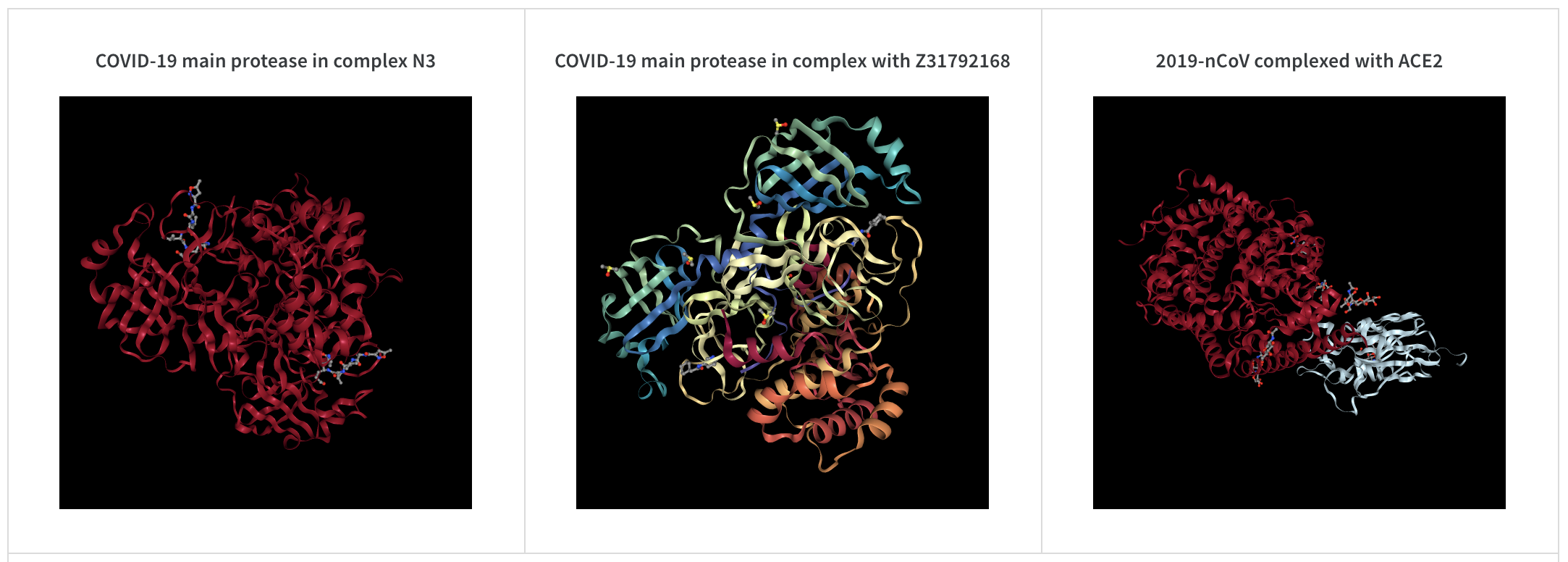
wandb.log({"protein": wandb.Molecule("6lu7.pdb")})
Log molecular data in any of 10 file types:pdb, pqr, mmcif, mcif, cif, sdf, sd, gro, mol2, or mmtf.
W&B also supports logging molecular data from SMILES strings, rdkit mol files, and rdkit.Chem.rdchem.Mol objects.
resveratrol = rdkit.Chem.MolFromSmiles("Oc1ccc(cc1)C=Cc1cc(O)cc(c1)O")
wandb.log(
{
"resveratrol": wandb.Molecule.from_rdkit(resveratrol),
"green fluorescent protein": wandb.Molecule.from_rdkit("2b3p.mol"),
"acetaminophen": wandb.Molecule.from_smiles("CC(=O)Nc1ccc(O)cc1"),
}
)
When your run finishes, you’ll be able to interact with 3D visualizations of your molecules in the UI.
See a live example using AlphaFold
PNG image
wandb.Image converts numpy arrays or instances of PILImage to PNGs by default.
wandb.log({"example": wandb.Image(...)})
# Or multiple images
wandb.log({"example": [wandb.Image(...) for img in images]})
Video
Videos are logged using the wandb.Video data type:
wandb.log({"example": wandb.Video("myvideo.mp4")})
Now you can view videos in the media browser. Go to your project workspace, run workspace, or report and click Add visualization to add a rich media panel.
2D view of a molecule
You can log a 2D view of a molecule using the wandb.Image data type and rdkit:
molecule = rdkit.Chem.MolFromSmiles("CC(=O)O")
rdkit.Chem.AllChem.Compute2DCoords(molecule)
rdkit.Chem.AllChem.GenerateDepictionMatching2DStructure(molecule, molecule)
pil_image = rdkit.Chem.Draw.MolToImage(molecule, size=(300, 300))
wandb.log({"acetic_acid": wandb.Image(pil_image)})
Other media
W&B also supports logging of a variety of other media types.
Audio
wandb.log({"whale songs": wandb.Audio(np_array, caption="OooOoo", sample_rate=32)})
A maximum of 100 audio clips can be logged per step. For more usage information, see audio-file.
Video
wandb.log({"video": wandb.Video(numpy_array_or_path_to_video, fps=4, format="gif")})
If a numpy array is supplied we assume the dimensions are, in order: time, channels, width, height. By default we create a 4 fps gif image (ffmpeg and the moviepy python library are required when passing numpy objects). Supported formats are "gif", "mp4", "webm", and "ogg". If you pass a string to wandb.Video we assert the file exists and is a supported format before uploading to wandb. Passing a BytesIO object will create a temporary file with the specified format as the extension.
On the W&B Run and Project Pages, you will see your videos in the Media section.
For more usage information, see video-file.
Text
Use wandb.Table to log text in tables to show up in the UI. By default, the column headers are ["Input", "Output", "Expected"]. To ensure optimal UI performance, the default maximum number of rows is set to 10,000. However, users can explicitly override the maximum with wandb.Table.MAX_ROWS = {DESIRED_MAX}.
columns = ["Text", "Predicted Sentiment", "True Sentiment"]
# Method 1
data = [["I love my phone", "1", "1"], ["My phone sucks", "0", "-1"]]
table = wandb.Table(data=data, columns=columns)
wandb.log({"examples": table})
# Method 2
table = wandb.Table(columns=columns)
table.add_data("I love my phone", "1", "1")
table.add_data("My phone sucks", "0", "-1")
wandb.log({"examples": table})
You can also pass a pandas DataFrame object.
table = wandb.Table(dataframe=my_dataframe)
For more usage information, see string.
HTML
wandb.log({"custom_file": wandb.Html(open("some.html"))})
wandb.log({"custom_string": wandb.Html('<a href="https://mysite">Link</a>')})
Custom HTML can be logged at any key, and this exposes an HTML panel on the run page. By default, we inject default styles; you can turn off default styles by passing inject=False.
wandb.log({"custom_file": wandb.Html(open("some.html"), inject=False)})
For more usage information, see html-file.
Feedback
Was this page helpful?
Glad to hear it! If you have more to say, please let us know.
Sorry to hear that. Please tell us how we can improve.